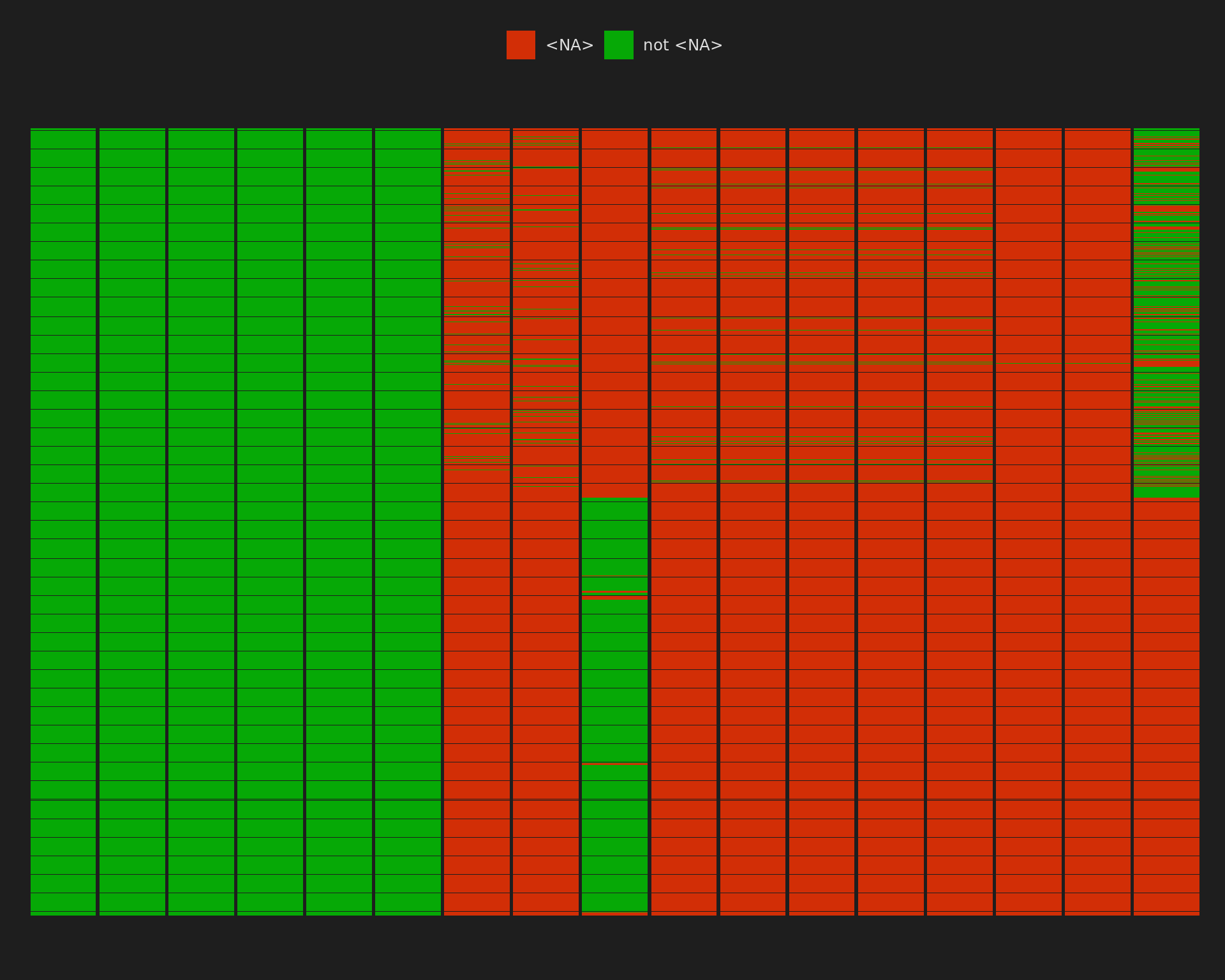
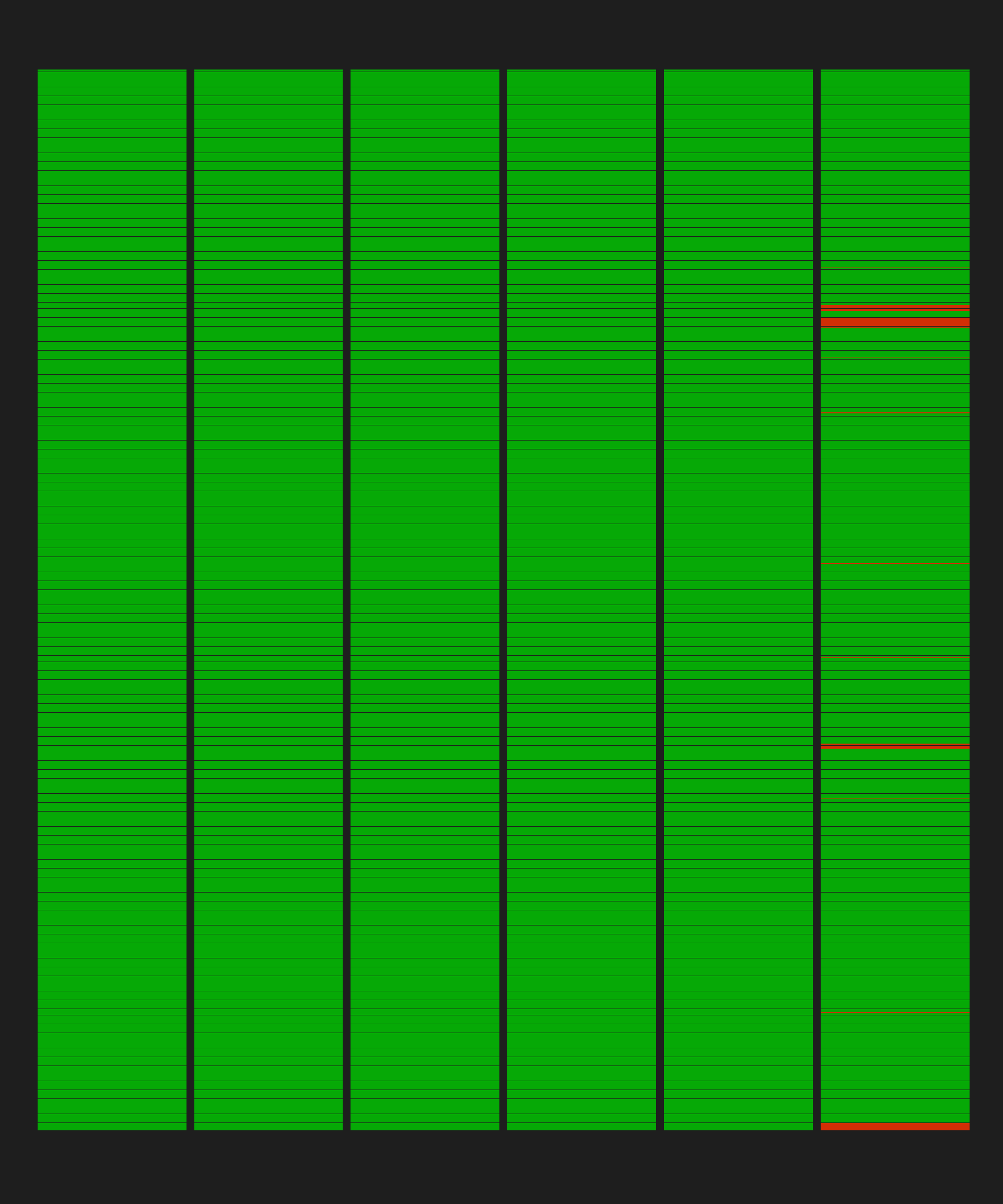
Rows: 9,236
Columns: 51
$ Index <dbl> 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10…
$ Date <date> 2022-02-17, 2022-02-17, 2022-02…
$ Time <time> 11:07:38, 11:02:39, 11:00:00, 1…
$ `New Device Time` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `BG Source` <chr> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `BG Reading (mg/dL)` <dbl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Linked BG Meter ID` <chr> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Basal Rate (U/h)` <dbl> NA, NA, 0.65, NA, NA, NA, NA, NA…
$ `Temp Basal Amount` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Temp Basal Type` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Temp Basal Duration (h:mm:ss)` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Bolus Type` <chr> "Normal", "Normal", NA, "Normal"…
$ `Bolus Volume Selected (U)` <dbl> 0.03, 0.05, NA, 0.05, 0.03, 0.03…
$ `Bolus Volume Delivered (U)` <dbl> 0.03, 0.05, NA, 0.05, 0.03, 0.03…
$ `Bolus Duration (h:mm:ss)` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Prime Type` <chr> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Prime Volume Delivered (U)` <dbl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ Alarm <chr> NA, NA, NA, NA, NA, NA, NA, NA, …
$ Suspend <chr> NA, NA, NA, NA, NA, NA, NA, NA, …
$ Rewind <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `BWZ Estimate (U)` <dbl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `BWZ Target High BG (mg/dL)` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `BWZ Target Low BG (mg/dL)` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `BWZ Carb Ratio (g/U)` <dbl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `BWZ Insulin Sensitivity (mg/dL/U)` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `BWZ Carb Input (grams)` <dbl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `BWZ BG Input (mg/dL)` <dbl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `BWZ Correction Estimate (U)` <dbl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `BWZ Food Estimate (U)` <dbl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `BWZ Active Insulin (U)` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `BWZ Status` <chr> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Sensor Calibration BG (mg/dL)` <dbl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Sensor Glucose (mg/dL)` <dbl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `ISIG Value` <dbl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Event Marker` <chr> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Bolus Number` <dbl> 236, 235, NA, 234, 233, 232, 231…
$ `Bolus Cancellation Reason` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `BWZ Unabsorbed Insulin Total (U)` <dbl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Final Bolus Estimate` <dbl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Scroll Step Size` <chr> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Insulin Action Curve Time` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Sensor Calibration Rejected Reason` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Preset Bolus` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Bolus Source` <chr> "CLOSED_LOOP_MICRO_BOLUS", "CLOS…
$ `BLE Network Device` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Network Device Associated Reason` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Network Device Disassociated Reason` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Network Device Disconnected Reason` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Sensor Exception` <chr> NA, NA, NA, NA, NA, NA, NA, NA, …
$ `Preset Temp Basal Name` <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …
$ ...51 <lgl> NA, NA, NA, NA, NA, NA, NA, NA, …